Timeseries
Timeseries transformations, filters, etc.
Classes:
|
Simple wrapper around |
|
Single gammatone filter based on [Sla97] |
|
Kalman filter!!!!! |
|
- class Filter_IIR(ftype='butter', buffer_size=256, coef_type='sos', axis=0, *args, **kwargs)[source]
Bases:
autopilot.transform.transforms.TransformSimple wrapper around
scipy.signal.iirfilter()Creates a streaming filter – takes in single values, stores them, and uses them to filter future values.
- Parameters
ftype (str) – filter type, see
ftypeofscipy.signal.iirfilter()for available filtersbuffer_size (int) – number of samples to store when filtering
coef_type ({‘ba’, ‘sos’}) – type of filter coefficients to use (see
scipy.signal.sosfilt()andscipy.signal.lfilt())axis (int) – which axis to filter over? (default: 0 because when passing arrays to filter, want to filter samples over time)
**kwargs – passed on to
scipy.signal.iirfilter(), eg.N- filter orderWn- array or scalar giving critical frequenciesbtype- type of band:['bandpass', 'lowpass', 'highpass', 'bandstop']
- Variables
coefs (np.ndarray) – filter coefficients, depending on
coef_typebuffer (collections.deque) – buffer of stored values to filter
coef_type (str) – type of filter coefficients to use (see
scipy.signal.sosfilt()andscipy.signal.lfilt())axis (int) – which axis to filter over? (default: 0 because when passing arrays to filter, want to filter samples over time)
ftype (str) – filter type, see
ftypeofscipy.signal.iirfilter()for available filters
Methods:
process(input)Filter the new value based on the values stored in
Filter.buffer
- class Gammatone(freq: float, fs: int, ftype: str = 'iir', filtfilt: bool = True, order: Optional[int] = None, numtaps: Optional[int] = None, axis: int = - 1, **kwargs)[source]
Bases:
autopilot.transform.transforms.TransformSingle gammatone filter based on [Sla97]
Thin wrapper around
scipy.signal.gammatone!! (started rewriting this and realized they had made a legible version <3 ty scipy team, additional implementations in the references)Examples
(Source code, png, hires.png, pdf)
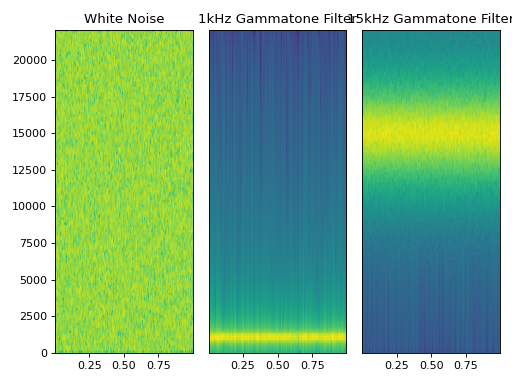
References
- Parameters
freq (float) – Center frequency of the filter in Hz
fs (int) – Sampling rate of the signal to process
ftype (str) – Type of filter to return from
scipy.signal.gammatone()filtfilt (bool) – If
True(default), usescipy.signal.filtfilt(), else usescipy.signal.lfilt()order (int) – From scipy docs: The order of the filter. Only used when
ftype='fir'. Default is 4 to model the human auditory system. Must be between 0 and 24.numtaps (int) – From scipy docs: Length of the filter. Only used when
ftype='fir'. Default isfs*0.015if fs is greater than 1000, 15 if fs is less than or equal to 1000.axis (int) – Axis of input signal to apply filter over (default
-1)**kwargs – passed to
scipy.signal.filtfilt()orscipy.signal.lfilt()
Methods:
process(input)
- class Kalman(dim_state: int, dim_measurement: Optional[int] = None, dim_control: int = 0, *args, **kwargs)[source]
Bases:
autopilot.transform.transforms.TransformKalman filter!!!!!
Adapted from https://github.com/rlabbe/filterpy/blob/master/filterpy/kalman/kalman_filter.py simplified and optimized lovingly <3
Each of the arrays is named with its canonical letter and a short description, (eg. the x_state vector
x_stateisself.x_state- Parameters
dim_state (int) – Dimensions of the state vector
dim_measurement (int) – Dimensions of the measurement vector
dim_control (int) – Dimensions of the control vector
- Variables
x_state (
numpy.ndarray) – Current state vectorP_cov (
numpy.ndarray) – Uncertainty CovarianceQ_proc_var (
numpy.ndarray) – Process UncertaintyB_control (
numpy.ndarray) – Control transition matrixF_state_trans (
numpy.ndarray) – State transition matrixH_measure (
numpy.ndarray) – Measurement functionR_measure_var (
numpy.ndarray) – Measurement uncertaintyM_proc_measure_xcor (
numpy.ndarray) – process-measurement cross correlationz_measure (
numpy.ndarray) –K (
numpy.ndarray) – Kalman gainy (
numpy.ndarray) –S (
numpy.ndarray) – System uncertaintySI (
numpy.ndarray) – Inverse system uncertaintyx_prior (
numpy.ndarray) – State priorP_prior (
numpy.ndarray) – Uncertainty priorx_post (
numpy.ndarray) – State posterior probabilityP_post (
numpy.ndarray) – Uncertainty posterior probability
References
Roger Labbe. “Kalman and Bayesian Filters in Python” - https://github.com/rlabbe/Kalman-and-Bayesian-Filters-in-Python Roger Labbe. “FilterPy” - https://github.com/rlabbe/filterpy
Methods:
predict([u, B, F, Q])Predict next x_state (prior) using the Kalman filter x_state propagation equations.
update(z[, R, H])Add a new measurement (z_measure) to the Kalman filter.
process(z, **kwargs)Call predict and update, passing the relevant kwargs
residual_of(z)Returns the residual for the given measurement (z_measure).
Helper function that converts a x_state into a measurement.
Attributes:
Fading memory setting.
- predict(u=None, B=None, F=None, Q=None)[source]
Predict next x_state (prior) using the Kalman filter x_state propagation equations.
Update our state and uncertainty priors,
x_priorandP_prior- unp.array, default 0
Optional control vector.
- Bnp.array(dim_state, dim_u), or None
Optional control transition matrix; a value of None will cause the filter to use self.B_control.
- Fnp.array(dim_state, dim_state), or None
Optional x_state transition matrix; a value of None will cause the filter to use self.F_state_trans.
- Qnp.array(dim_state, dim_state), scalar, or None
Optional process noise matrix; a value of None will cause the filter to use self.Q_proc_var.
- update(z: numpy.ndarray, R=None, H=None) numpy.ndarray[source]
Add a new measurement (z_measure) to the Kalman filter.
If z_measure is None, nothing is computed. However, x_post and P_post are updated with the prior (x_prior, P_prior), and self.z_measure is set to None.
- Parameters
z (
numpy.ndarray) – measurement for this update. z_measure can be a scalar if dim_measurement is 1, otherwise it must be convertible to a column vector.If you pass in a value of H_measure, z_measure must be a column vector the of the correct size.
R (
numpy.ndarray, int, None) – Optionally provide R_measure_var to override the measurement noise for this one call, otherwise self.R_measure_var will be used.H (
numpy.ndarray, None) – Optionally provide H_measure to override the measurement function for this one call, otherwise self.H_measure will be used.
- process(z, **kwargs)[source]
Call predict and update, passing the relevant kwargs
- Parameters
z ()
**kwargs ()
- Returns
self.x_state
- Return type
np.ndarray
- residual_of(z)[source]
Returns the residual for the given measurement (z_measure). Does not alter the x_state of the filter.
- measurement_of_state(x)[source]
Helper function that converts a x_state into a measurement.
- xnp.array
kalman x_state vector
- z_measure(dim_measurement, 1): array_like
measurement for this update. z_measure can be a scalar if dim_measurement is 1, otherwise it must be convertible to a column vector.
- property alpha
Fading memory setting. 1.0 gives the normal Kalman filter, and values slightly larger than 1.0 (such as 1.02) give a fading memory effect - previous measurements have less influence on the filter’s estimates. This formulation of the Fading memory filter (there are many) is due to Dan Simon [1]_.